Real-Time Shortest Paths on Dynamic Networks with Bellman-Ford in Pathway
 Pathway Team
Pathway TeamThis article explains step-by-step how the Bellman–Ford algorithm may be implemented in Pathway.
Introduction
The Bellman-Ford algorithm computes the shortest paths from a single source vertex to all the other vertices in a weighted graph. A weighted graph is composed of a set of points, called vertices, which are connected via edges. Each edge is associated to a value, called either weight or distance. For instance, the set of all the cities and the roads which connect them form such a graph. In that example, the Bellman-Ford algorithm would help to find the fastest way, in terms of distance, to go from a given city to another.
This article is also a perfect place to familiarize yourself with several constructs used in Pathway.
Code
First things first - imports 🙂
import math
import pathway as pw
I/O Data
The input is a weighted graph so it is natural to split representation of the data into two parts: Vertices and Edges. Their schemas:
class Vertex(pw.Schema):
is_source: bool
class Edge(pw.Schema):
u: pw.Pointer[Vertex]
v: pw.Pointer[Vertex]
dist: int
These schemas have a natural interpretation. You can think of the Edge schema as of
a blueprint of a table that has 3 columns: u, v (foreign keys) and dist.
The output schema:
class DistFromSource(pw.Schema):
dist_from_source: int
Note: The schemas inherit from pw.Schema special class.
Note: You might wonder why output schema has only one column dist_from_source.
Actually, you can join schemas together to create a new one. And so, the output schema
is Vertex + DistFromSource. (Look for that type annotation later in the code.)
The algorithm
The Bellman-Ford algorithm performs some number of relaxations until it reaches a fixed point.
Relaxations
Each node checks if a path via it would make any so-far-optimal path to some other node shorter.
def bellman_ford_step(
vertices_dist: pw.Table[DistFromSource], edges: pw.Table[Edge]
) -> pw.Table[DistFromSource]:
relaxed_edges = edges + edges.select(
dist_from_source=vertices_dist.ix(edges.u).dist_from_source + edges.dist
)
vertices_dist = vertices_dist.update_rows(
relaxed_edges.groupby(id=relaxed_edges.v).reduce(
dist_from_source=pw.reducers.min(relaxed_edges.dist_from_source),
)
)
return vertices_dist
Fixed point
The relaxations are iterated until a fixed point is reached. In this case, reaching a fixed point means that no new (shorter) path was found in the last iteration.
def bellman_ford(vertices: pw.Table[Vertex], edges: pw.Table[Edge]):
vertices_dist: pw.Table[DistFromSource] = vertices.select(
dist_from_source=pw.if_else(vertices.is_source, 0.0, math.inf)
)
fixed_point = pw.iterate(
lambda iterated, edges: dict(
iterated=bellman_ford_step(vertices_dist=iterated, edges=edges)
),
# The `pw.iterate_universe` stanza informs iterate that `vertices_dist` grows with each loop iteration. Without it, the system assumes that iterations don't change the set of indices of a table.
iterated=pw.iterate_universe(vertices_dist),
edges=edges,
).iterated
return fixed_point.join(vertices, fixed_point.id == vertices.id).select(
vertices.key, fixed_point.dist_from_source
)
Tests
Now, let's see the code in action. The following test case runs Bellman-Ford algorithm on a graph depicted below.
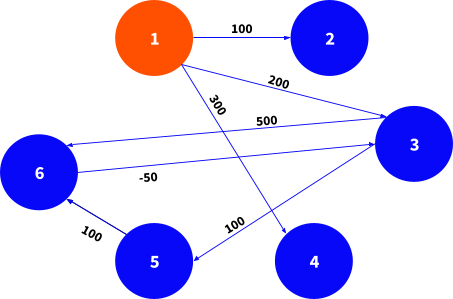
# a directed graph
vertices = pw.debug.table_from_markdown(
"""
| key | is_source
1 | 1 | True
2 | 2 | False
3 | 3 | False
4 | 4 | False
5 | 5 | False
6 | 6 | False
7 | 7 | False
"""
).with_id_from(pw.this.key)
edges = pw.debug.table_from_markdown(
"""
| u | v | dist
11 | 1 | 2 | 100
12 | 1 | 3 | 200
13 | 1 | 4 | 300
14 | 3 | 5 | 100
15 | 3 | 6 | 500
16 | 5 | 6 | 100
17 | 6 | 3 | -50
"""
).with_columns(
u=vertices.pointer_from(pw.this.u),
v=vertices.pointer_from(pw.this.v),
)
Pathway automatically reindexes the tables, so we need a key column of the vertices table and we need ask Pathway to reindex the table using those.
In practice, Pathway uses pointers so the keys are automatically converted into pointers.
For the edges, we have to convert the keys into their references in order to be able to use vertices_dist.ix(edges.u) as ix only works with pointers.
pw.debug.compute_and_print(bellman_ford(vertices, edges))
| key | dist_from_source
^YKEMVCM... | 1 | 0.0
^MDY4T58... | 2 | 100.0
^3AYGT7R... | 3 | 200.0
^FGFPAGV... | 4 | 300.0
^81GRBTV... | 5 | 300.0
^P358CHY... | 6 | 400.0
^CTWGSGG... | 7 | inf
That was a simple introduction to writing code and tests in Pathway.
Feel free to take this code and experiment with it 🙂 Do you see any possibility to improve the code? (What happens when there is a negative cycle in the graph?)
Summary
The code above follows a pattern that is quite frequent when working with Pathway:
- Define I/O data types
- Define transformations on tables
- Iterate the transformation until a fixed point is reached
- usually transforms the data by a simple one-liner.
- for example
iterate(lambda foo, bar: {foo=fn(foo, bar), bar=bar}, foo=input_table_1, bar=input_table2).foo

Pathway Team
 Olivier Ruas
Olivier Ruas tutorial · machine-learningOct 26, 2022Realtime Classification with Nearest Neighbors
tutorial · machine-learningOct 26, 2022Realtime Classification with Nearest Neighbors Saksham Goel
Saksham Goel blog · tutorialFeb 5, 2025Real-Time AI Pipeline with DeepSeek, Ollama and Pathway
blog · tutorialFeb 5, 2025Real-Time AI Pipeline with DeepSeek, Ollama and Pathway Shlok Srivastava
Shlok Srivastava tutorialDec 11, 2024Scalable Alternative to Apache Kafka and Flink for Advanced Streaming: Build Real-Time Systems with NATS and Pathway
tutorialDec 11, 2024Scalable Alternative to Apache Kafka and Flink for Advanced Streaming: Build Real-Time Systems with NATS and Pathway
