Building a Real-Time Radiology AI System with Pathway and LandingAI
 Ishan Upadhyay
Ishan UpadhyayHealthcare teams process thousands of radiology reports every day, each packed with important diagnostic information in the form of images and text. When you can analyze these documents in real-time, it opens up possibilities for much faster information access and better clinical workflows.
I've been working on a system that brings together LandingAI's intelligent document processing capabilities with Pathway's real-time server architecture to tackle this exact challenge. Underneath it leverages the DPT-2 architecture recently released by Andrew Ng. My idea is straightforward: as radiology reports come in as PDFs, the system accurately pulls out the key medical findings, organizes them, and makes everything instantly searchable for healthcare teams. This means doctors can quickly find what they need from complex medical reports without having to dig through pages of unstructured text. The system handles the heavy lifting of document processing while healthcare professionals focus on what they do best.
Project Repository: RadiologyAI on GitHub
LandingAI Parser UDF
The core document processing engine leverages LandingAI's Agentic Document Extraction (ADE) API to parse complex radiology report PDFs and extract unstructured medical information. This is done with the help of a custom User Defined Function (UDF) in Pathway, that handles the heavy lifting of converting unstructured medical narratives into searchable, actionable data on the go. ADE (my choice of parser here) extracts the contents of the file in raw markdown as well as a structured schema from PDF based radiology reports, making them instantly available for real-time analysis and alerting via real-time indexing, which is easy to implement with Pathway.
Getting Started: You'll need a LandingAI API key and some credits for testing the parsing (you get $10 free credit on first sign up).
Defining the Extraction Schema
For extracting document information based on a structured schema, define a Python dictionary object that specifies the schema for data extraction:
extraction_schema = {
"type": "object",
"properties": {
"patient_id": {
"type": "string",
"description": "Patient identification number or ID"
},
"study_type": {
"type": "string",
"description": "Type of radiological study (CT, MRI, X-ray, Ultrasound, etc.)"
},
"findings": {
"type": "string",
"description": "Key radiological findings and observations from the study"
},
"impression": {
"type": "string",
"description": "Radiologist's impression, conclusion, and clinical interpretation"
},
"critical_findings": {
"type": "string",
"description": "Any critical, urgent, or life-threatening findings requiring immediate attention"
}
},
"additionalProperties": False,
"required": ["study_type", "findings", "impression"]
}
LandingAI Parser Implementation
Here's my implementation of the LandingAI parser as a Pathway UDF:
import pathway as pw
from typing import List, Optional
import logging
from pathlib import Path
from pydantic import BaseModel, Field
from agentic_doc.parse import parse
from agentic_doc.config import ParseConfig
logger = logging.getLogger(__name__)
class LandingAIRadiologyParser(pw.UDF):
"""Parse radiology reports using LandingAI agentic-doc library."""
def __init__(self, api_key: str, capacity: int, results_dir: str, cache_strategy: pw.udfs.CacheStrategy = None, *, async_mode: str = "fully_async", **kwargs):
self.api_key = api_key
self.async_mode = async_mode
self.results_dir = results_dir
self.capacity = capacity
from pathway.xpacks.llm._utils import _prepare_executor
executor = _prepare_executor(async_mode)
super().__init__(cache_strategy=cache_strategy, executor=executor)
async def parse(self, contents: bytes) -> List[tuple[str, dict]]:
"""Parse radiology reports using LandingAI."""
results_dir = Path(self.results_dir)
results_dir.mkdir(exist_ok=True)
# Define extraction schema in JSON Schema format
extraction_schema = {
"type": "object",
"properties": {
"patient_id": {
"type": "string",
"description": "Patient identification number or ID"
},
"study_type": {
"type": "string",
"description": "Type of radiological study (CT, MRI, X-ray, Ultrasound, etc.)"
},
"findings": {
"type": "string",
"description": "Key radiological findings and observations from the study"
},
"impression": {
"type": "string",
"description": "Radiologist's impression, conclusion, and clinical interpretation"
},
"critical_findings": {
"type": "string",
"description": "Any critical, urgent, or life-threatening findings requiring immediate attention"
}
},
"additionalProperties": False,
"required": ["study_type", "findings", "impression"]
}
# Parse document with LandingAI using proper JSON Schema
parsed_results = parse(
contents,
include_marginalia=True,
include_metadata_in_markdown=True,
result_save_dir=str(results_dir),
extraction_schema=extraction_schema,
config=ParseConfig(api_key=self.api_key)
)
if not parsed_results:
return [("", {"source": "landingai", "error": "No parsing results"})]
parsed_doc = parsed_results[0]
text_content = getattr(parsed_doc, 'markdown', "")
# Extract structured data from extraction_metadata if available
extraction_data = {}
if hasattr(parsed_doc, 'extraction_metadata') and parsed_doc.extraction_metadata:
for field, data in parsed_doc.extraction_metadata.items():
if isinstance(data, dict) and 'value' in data and data['value']:
extraction_data[field] = data['value']
# Create clean metadata with extracted fields
metadata = {
"source": "landingai",
"confidence": str(getattr(parsed_doc, 'confidence', 0.0)),
**{k: str(v) for k, v in extraction_data.items() if v is not None}
}
# Ensure string types for Pathway
safe_text = str(text_content) if text_content else ""
safe_metadata = {k: str(v) if v is not None else "" for k, v in metadata.items()}
return [(safe_text, safe_metadata)]
async def __wrapped__(self, contents: bytes, **kwargs) -> list[tuple[str, dict]]:
return await self.parse(contents)
Configuration Setup
Great! Now half of the work is done. Let's update our app.yaml config to use the LandingAI parser:
Note: If you want to use OpenAI API for token embedding and model calling then uncomment the relevant lines.
$parser: !src.parsers.landingai_parser.LandingAIRadiologyParser
api_key: $LANDINGAI_API_KEY
async_mode: "fully_async"
Configuration Parameters:
async_mode: Execution mode for the UDF
MCP Server Configuration
For running an MCP server, add the following configuration to app.yaml:
mcp_server: !pw.xpacks.llm.mcp_server.PathwayMcp
name: "CriticalAlert AI MCP Server"
transport: "streamable-http"
host: "localhost"
port: 8123
serve:
- $document_store
Complete app.yaml Configuration
path: data/incoming
format: binary
with_metadata: true
mode: streaming
$llm: !pw.xpacks.llm.llms.LiteLLMChat
model: "anthropic/claude-3-5-sonnet-20241022"
api_key: $ANTHROPIC_API_KEY
retry_strategy: !pw.udfs.ExponentialBackoffRetryStrategy
max_retries: 6
cache_strategy: !pw.udfs.DefaultCache {}
temperature: 0
capacity: 8
$embedder: !pw.xpacks.llm.embedders.SentenceTransformerEmbedder
model: "all-MiniLM-L12-v2"
$splitter: !pw.xpacks.llm.splitters.TokenCountSplitter
max_tokens: 800
min_tokens: 200
$parser: !src.parsers.landingai_parser.LandingAIRadiologyParser
api_key: $LANDINGAI_API_KEY
async_mode: "batch_async"
capacity: 4
$retriever_factory: !pw.stdlib.indexing.BruteForceKnnFactory
reserved_space: 1000
embedder: $embedder
metric: !pw.stdlib.indexing.BruteForceKnnMetricKind.COS
$document_store: !src.parsers.landingai_parser.RadiologyDocumentStore
docs: $sources
retriever_factory: $retriever_factory
splitter: $splitter
landingai_api_key: $LANDINGAI_API_KEY
# cache_strategy removed to prevent Pydantic serialization issues
question_answerer: !pw.xpacks.llm.question_answering.BaseRAGQuestionAnswerer
llm: $llm
indexer: $document_store
search_topk: 6
mcp_server: !pw.xpacks.llm.mcp_server.PathwayMcp
name: "Radiology MCP Server"
transport: "streamable-http"
host: "localhost"
port: 8123
serve:
- $document_store
host: "0.0.0.0"
port: 49001
with_cache: true
terminate_on_error: false
Main Application
Now let's write the main file that will run the real-time parsing + indexing pipeline along with REST and MCP servers running.
import logging
import os
import pathway as pw
from dotenv import load_dotenv
from pydantic import BaseModel, ConfigDict, InstanceOf
from pathway.xpacks.llm.servers import QASummaryRestServer
from pathway.xpacks.llm.question_answering import BaseRAGQuestionAnswerer
from pathway.xpacks.llm.mcp_server import PathwayMcp
logging.basicConfig(
level=logging.INFO,
format="%(asctime)s %(name)s %(levelname)s %(message)s",
datefmt="%Y-%m-%d %H:%M:%S",
)
load_dotenv()
pw.set_license_key(os.getenv("PATHWAY_LICENSE_KEY"))
def _env_flag(name: str, default: bool = False) -> bool:
value = os.getenv(name)
if value is None:
return default
return value.lower() in {"1", "true", "yes", "on"}
DEBUG_UPDATE_STREAM = _env_flag("PW_DEBUG_UPDATE_STREAM", False)
class App(BaseModel):
"""
CriticalAlert AI Application
Follows the exact same pattern as question_answering_rag/app.py
The YAML instantiates the critical_alert_answerer which contains the RadiologyDocumentStore
"""
question_answerer: InstanceOf[BaseRAGQuestionAnswerer]
mcp_server: InstanceOf[PathwayMcp] = None
# Server configuration
host: str
port: int
with_cache: bool = True
terminate_on_error: bool = False
debug_update_stream: bool = DEBUG_UPDATE_STREAM
def run(self) -> None:
"""
Run the CriticalAlert AI application
This is where the actual Pathway computation graph gets set up
Similar to QASummaryRestServer.run() in existing examples
"""
server = QASummaryRestServer(self.host, self.port, self.question_answerer)
if self.mcp_server:
logging.info(f"MCP Server: http://{self.mcp_server.host}:{self.mcp_server.port}/mcp/")
server.run(
with_cache=self.with_cache,
terminate_on_error=self.terminate_on_error,
cache_backend=pw.persistence.Backend.filesystem("Cache"),
)
@classmethod
def from_config(cls, config: dict) -> "App":
"""Instantiate App from YAML config while honoring debug overrides."""
config = dict(config)
debug_override = config.pop("debug_update_stream", None)
instance = cls(**config)
if debug_override is not None:
instance.debug_update_stream = bool(debug_override)
return instance
model_config = ConfigDict(extra="forbid")
if __name__ == "__main__":
with open("app.yaml") as f:
config = pw.load_yaml(f)
app = App.from_config(config)
app.run()
Key Pathway Components
You'll notice we use two main Pathway classes:
BaseRAGQuestionAnswerer: The core RAG (Retrieval-Augmented Generation) engine that combines document retrieval with LLM generation. It searches through your indexed documents to find relevant context, then uses an LLM to generate accurate answers based on that retrieved information. You can customize this class to implement domain-specific retrieval strategies, modify the prompt templates, or add custom post-processing logic for their specific use case.
QASummaryRestServer: A REST API server that exposes the RAG question-answering capabilities through HTTP endpoints like /v1/retrieve and /v1/answer. It handles incoming queries, coordinates with the question answerer, and returns structured JSON responses. Users can customize this class to add authentication, modify API response formats, or create custom endpoints tailored to their application's needs.
Both classes work together to provide servable RAG that can be easily customized.
Running the Application
Now let's run our application - it's as simple as:
python app.py
Note: Make sure to set up the proper environment before running your app
Testing the System
For testing features, you can use REST methods or MCP clients like Claude, Cursor, etc.
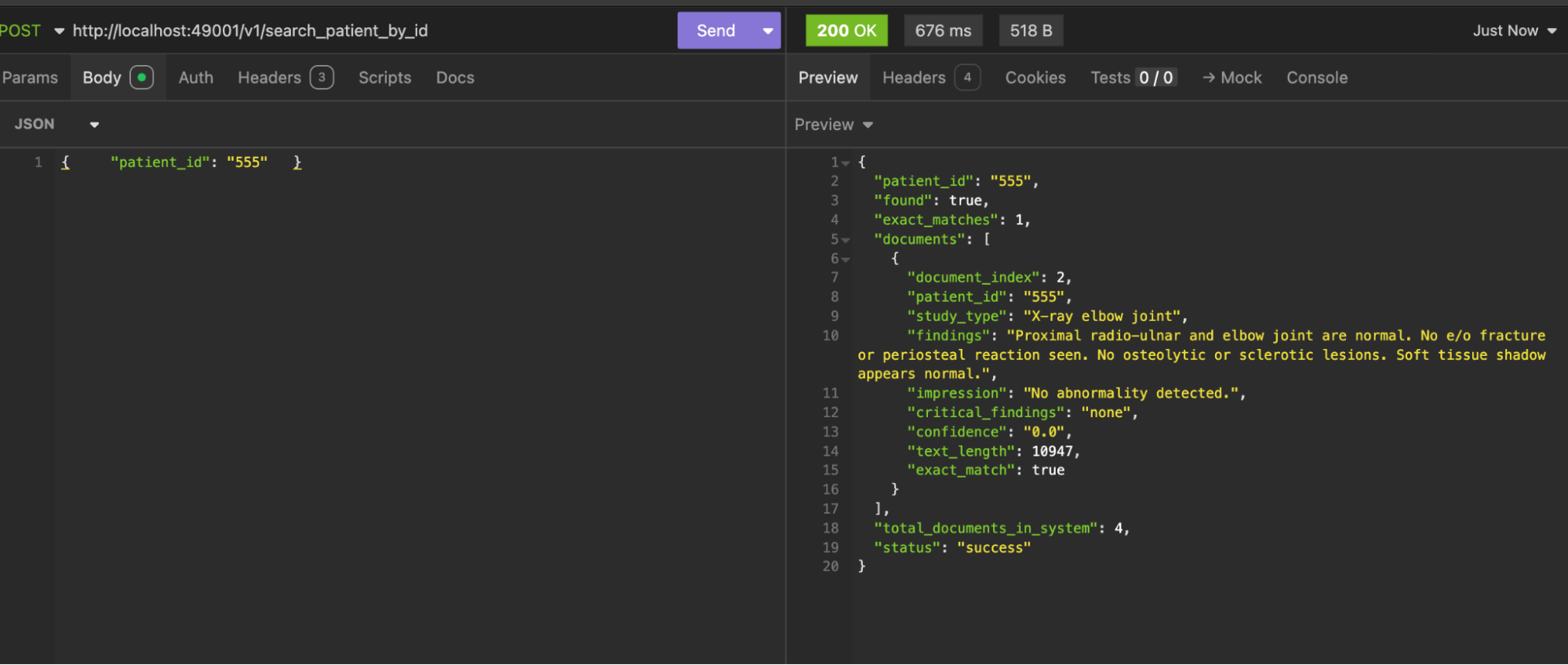
For all available API methods, check out the QARestServer documentation or visit the GitHub repository.
MCP Integration
Now that we've added the relevant settings for the MCP server in our config, you should see in the logs that an MCP server has started on localhost:8123 with all registered tools.
For Cursor, you can add this config to .cursor/mcp.json:
{
“mcpServers”: {
"critical-alert-ai": {
"url": "http://localhost:8123/mcp/"
}
}
}
You need to turn on your mcp server from the settings -> Tools and Integration and it should show what all tools your model can use particularly: retrieve_query, statistics_query, inputs_query.

Sounds easy, right?
Now let's move to a slightly more advanced part.
Remember the classes I mentioned: QAAnswerer and QARestServer. There's one more class you need to know: DocumentStore, which is responsible for automatically indexing and storing the documents it gets from the parser.
For my use case, I needed to expose extra tools and endpoints that can query based on patient_id.
Custom Document Store
Let's create a custom document store so we can expose UDFs as MCP-servable tools on the same endpoint we configured before.
When deriving the DocumentStore, add the following method:
class PatientSearchSchema(pw.Schema):
"""Schema for patient search by ID"""
patient_id: str = pw.column_definition(dtype=str, default_value="")
class PatientQueryResultSchema(pw.Schema):
"""Result schema for patient extraction query - matches DocumentStore pattern"""
result: pw.Json
@pw.table_transformer
def search_patient_by_id(self, request_table: pw.Table[PatientSearchSchema]) -> pw.Table[PatientSearchResultSchema]:
"""
MCP Tool: Search for specific patient ID in parsed documents.
"""
logger.info("🔍 search_patient_by_id: Filtering by specific patient ID")
all_docs = self.parsed_docs.reduce(
metadatas=pw.reducers.tuple(pw.this.metadata),
texts=pw.reducers.tuple(pw.this.text),
total_docs=pw.reducers.count(),
)
@pw.udf
def search_by_patient_id(requested_patient_id: str, metadatas: list, texts: list, total_docs: int) -> pw.Json:
matching_docs = []
for i, (metadata, text) in enumerate(zip(metadatas or [], texts or [])):
if metadata:
try:
metadata_dict = metadata.as_dict() if hasattr(metadata, 'as_dict') else {}
doc_patient_id = str(metadata_dict.get("patient_id", "")).strip()
# Check if this document matches the requested patient ID
if doc_patient_id == requested_patient_id or (doc_patient_id and doc_patient_id != "unknown"):
matching_docs.append({
"document_index": i + 1,
"patient_id": doc_patient_id,
"study_type": metadata_dict.get("study_type", "unknown"),
"findings": str(metadata_dict.get("findings", ""))[:200] + "..." if len(str(metadata_dict.get("findings", ""))) > 200 else str(metadata_dict.get("findings", "")),
"impression": str(metadata_dict.get("impression", ""))[:200] + "..." if len(str(metadata_dict.get("impression", ""))) > 200 else str(metadata_dict.get("impression", "")),
"critical_findings": metadata_dict.get("critical_findings", "none"),
"confidence": metadata_dict.get("confidence", "0.0"),
"text_length": len(str(text)) if text else 0,
"exact_match": doc_patient_id == requested_patient_id
})
except:
continue
if matching_docs:
exact_matches = [doc for doc in matching_docs if doc["exact_match"]]
if exact_matches:
response = {
"patient_id": requested_patient_id,
"found": True,
"exact_matches": len(exact_matches),
"documents": exact_matches,
"total_documents_in_system": total_docs,
"status": "success"
}
else:
response = {
"patient_id": requested_patient_id,
"found": False,
"similar_patients": matching_docs,
"total_documents_in_system": total_docs,
"status": "no_exact_match",
"message": f"No exact match for patient ID '{requested_patient_id}', but found {len(matching_docs)} other patients"
}
else:
response = {
"patient_id": requested_patient_id,
"found": False,
"total_documents_in_system": total_docs,
"status": "not_found",
"message": f"No documents found for patient ID '{requested_patient_id}'"
}
return pw.Json(response)
result = request_table.join_left(all_docs, id=request_table.id).select(
result=search_by_patient_id(
request_table.patient_id,
all_docs.metadatas,
all_docs.texts,
all_docs.total_docs
)
)
return result
How It Works
This method queries the Pathway table with the patientId we provide, performs table operations using Pathway's UDF, and returns the required result in JSON format.
You can add more custom methods for querying the table for different use cases. Additionally, the data we get has second-level latency, which can be very useful in time-critical scenarios like healthcare.
Registering MCP Tools
Now we need to register this as a tool for our MCP server. For that, we need to override the register_mcp method:
def register_mcp(self, server):
"""
Register MCP tools including both DocumentStore defaults and custom patient tools.
This overrides the parent DocumentStore.register_mcp to add our custom patient tools
alongside the standard retrieve_query, statistics_query, and inputs_query tools.
"""
super().register_mcp(server)
server.tool(
name="search_patient_by_id",
request_handler=self.search_patient_by_id,
schema=self.PatientSearchSchema,
)
Great! Now we can actually use this MCP in our Cursor client and give it access to the live data that we're getting from the parser PDF documents.
Tip: You'll need to re-run
app.pyfor the effect to take place. If your MCP client errors out as "invalid session," restart it once.

REST Endpoints for Patient Queries
Great! Now we have custom tools exposed using the MCP server. Let's also expose a REST endpoint for querying based on patient_id.
For this, you need to create a derived class of BaseRAGQuestionAnswerer and add this method to it:
class PatientSearchSchema(pw.Schema):
patient_id: str = pw.column_definition(dtype=str, default_value="")
@pw.table_transformer
def search_patient_by_id(self, request_table: pw.Table[PatientSearchSchema]) -> pw.Table:
"""
Search for patient by ID - delegates to DocumentStore implementation.
"""
return self.indexer.search_patient_by_id(request_table)
Custom REST Server
Here's the final step before we can try out our custom endpoint. Create a derived class of QARestServer and override the __init__ method like this:
def __init__(
self,
host: str,
port: int,
rag_question_answerer: RadiologyQuestionAnswerer,
**rest_kwargs,
):
# QARestServer already registers all standard endpoints (retrieve, statistics, etc.)
super().__init__(host, port, rag_question_answerer, **rest_kwargs)
# Only register our custom patient-specific endpoints
self.serve(
"/v3/search_patient_by_id",
rag_question_answerer.PatientSearchSchema,
rag_question_answerer.search_patient_by_id,
**rest_kwargs,
)
Project Structure
Great! Now everything is set up. Keep your directory structure as follows:
├── app.py # Main application entry point
├── app.yaml # Application configuration
├── src/
│ ├── __init__.py # Package initialization
│ ├── parsers/
│ │ ├── __init__.py
│ │ └── landingai_parser.py # LandingAI document parser
│ ├── intelligence/
│ │ ├── __init__.py
│ │ └── critical_alert_answerer.py # RAG question answerer
│ ├── store/
│ │ └── RadiologyDocumentStore.py # Document store with MCP tools
│ └── server/
│ └── RadiologyServer.py # REST API server
├── data/
│ ├── incoming/ # Drop PDF files here
│ └── processed/ # Processed documents
└── Cache/ # Pathway cache directory
Final Configuration Update
Also update the app.yaml file to use the correct configuration:
$sources: !pw.io.fs.read
path: data/incoming
format: binary
with_metadata: true
mode: streaming
$llm: !pw.xpacks.llm.llms.LiteLLMChat
model: "anthropic/claude-3-5-sonnet-20241022"
api_key: $ANTHROPIC_API_KEY
retry_strategy: !pw.udfs.ExponentialBackoffRetryStrategy
max_retries: 6
cache_strategy: !pw.udfs.DefaultCache {}
temperature: 0
capacity: 8
$embedder: !pw.xpacks.llm.embedders.SentenceTransformerEmbedder
model: "all-MiniLM-L12-v2"
$splitter: !pw.xpacks.llm.splitters.TokenCountSplitter
max_tokens: 800
min_tokens: 200
$parser: !src.parsers.landingai_parser.LandingAIRadiologyParser
api_key: $LANDINGAI_API_KEY
capacity: 4
results_dir: "data/extraction_results"
cache_strategy: !pw.udfs.DefaultCache {}
async_mode: "fully_async"
$retriever_factory: !pw.stdlib.indexing.BruteForceKnnFactory
reserved_space: 1000
embedder: $embedder
metric: !pw.stdlib.indexing.BruteForceKnnMetricKind.COS
$document_store: !src.store.RadiologyDocumentStore.RadiologyDocumentStore
docs: $sources
retriever_factory: $retriever_factory
splitter: $splitter
parser: $parser
question_answerer: !src.intelligence.critical_alert_answerer.RadiologyQuestionAnswerer
llm: $llm
indexer: $document_store
search_topk: 6
mcp_server: !pw.xpacks.llm.mcp_server.PathwayMcp
name: "Document Processing MCP Server"
transport: "streamable-http"
host: "localhost"
port: $MCP_PORT
serve:
- $document_store
host: "0.0.0.0"
port: $REST_PORT
with_cache: true
terminate_on_error: false
Testing the Complete System
Great! Now let's test our app again by running:
python app.py
You should now be able to query using the API endpoint based on patient_id!
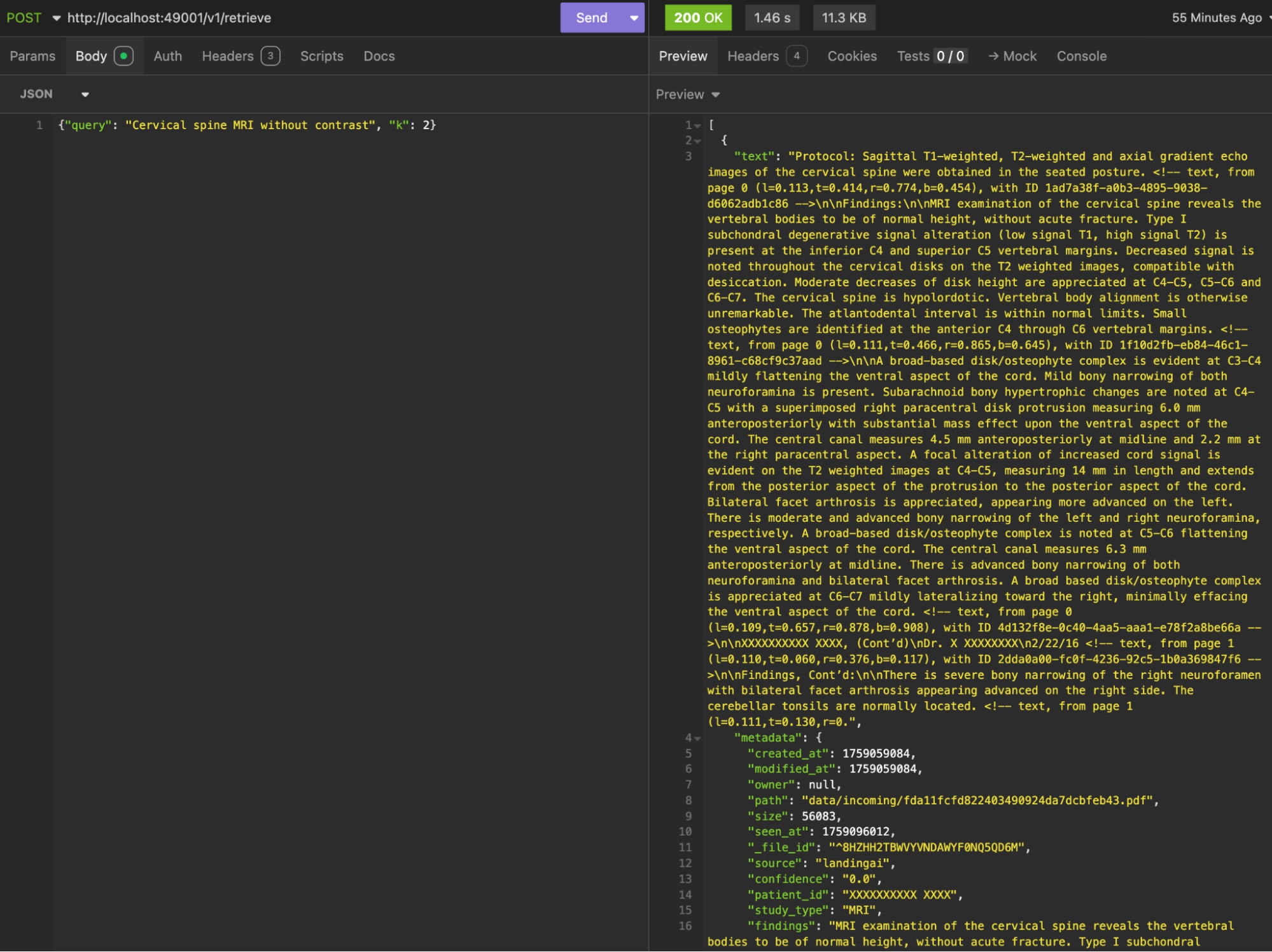
What We've Built
Now we have a complete pipeline running that:
- Constantly checks for new radiology reports in our directory
- Accurately Parses them using LandingAI ADE with structured extraction
- Makes the live indexes and metadata queryable in real-time
- Exposes custom endpoints for patient-specific queries
- Provides MCP integration for AI assistants like Cursor and Claude
Future Enhancements
I'd love to hear your thoughts on how we can make this better! Some ideas for additional custom endpoints:
- Critical alerts endpoint for urgent findings
- Study type analytics for radiological trends
- Real-time notifications for critical findings using websockets
To Support the Project
If you found this blog helpful, please consider:
- Starring the repositories: Pathway, LandingAI, and RadiologyAI
- Sharing this blog with developers who are working on time-critical AI workflows
 Ishan Upadhyay
Ishan Upadhyay communityOct 2, 2025Building a Real-Time Radiology AI System with Pathway and LandingAI
communityOct 2, 2025Building a Real-Time Radiology AI System with Pathway and LandingAI Shlok Srivastava
Shlok Srivastava communityFeb 28, 2025Exploring Kappa Architecture with Pathway
communityFeb 28, 2025Exploring Kappa Architecture with Pathway Pathway Community
Pathway Community communityApr 2, 2025Adaptive Agents for Real-Time RAG: Domain-Specific AI for Legal, Finance & Healthcare
communityApr 2, 2025Adaptive Agents for Real-Time RAG: Domain-Specific AI for Legal, Finance & Healthcare

