Pathway + PostgreSQL + LLM: app for querying financial reports with live document structuring pipeline.
 Berke Can Rizai
Berke Can RizaiPathway + PostgreSQL + LLM: app for querying financial reports with live document structuring pipeline.
This showcase demonstrates a data pipeline that calls into LLMs for document processing. In the showcase, you will see how Pathway can extract information from documents and keep the results up to date when documents change.
About the project
Unformatted raw text is everywhere: PDFs, Word documents, websites, emails, etc. Extracting meaningful information from this unformatted raw text is extremely valuable. In the past, the solution was to label data by hand and train custom models specializing in a specific task. This approach requires too many resources. However, with the advent of LLMs, we can now set up a project to structure unformatted raw text in minutes. Creating POC and verifying use cases is now particularly easy without spending too much time.
The following video shows document structurization in action as implemented in the unstructured_to_sql example of Pathway LLM-App.
The unstructured_to_sql app consists of two parts:
- Ingesting data from documents into PostgreSQL.
- Retrieving information from PostgreSQL with natural language.
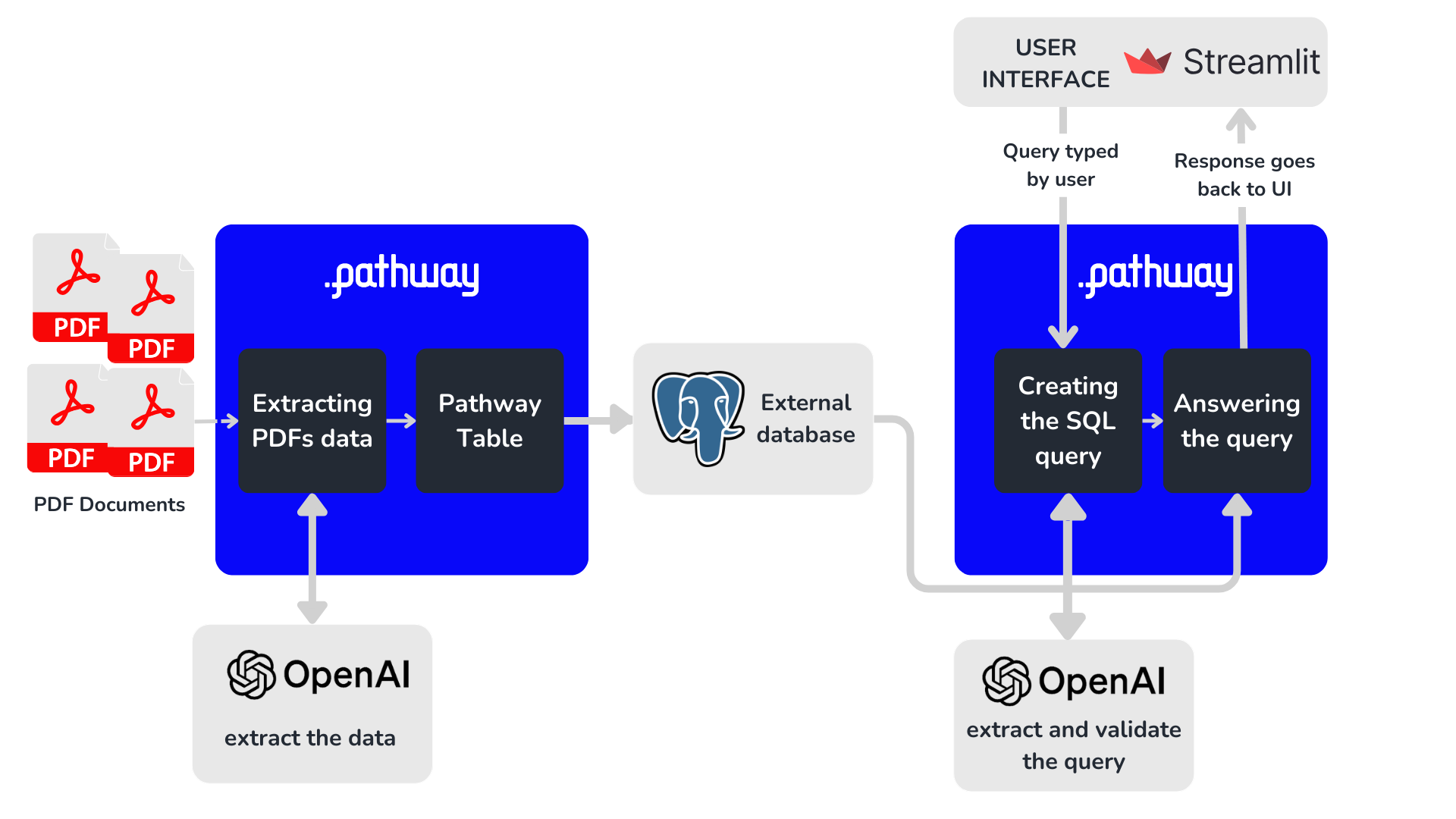
This showcase will focus on the ingestion of PDF content (part 1), which is more relevant for streaming data. In a nutshell, the ingestion process can be broken down into three separate steps:
- Reading text from PDFs.
- Extracting information desired by the user and creating a Pathway table.
- Inserting the Pathway table into PostgreSQL.
1. Reading text from PDFs
The first step is to read the PDFs and store the unformatted text in a Pathway table.
With Pathway, you can take input from any arbitrary connector. For this example, we will read PDFs from a local directory. This could also be:
- a Google Drive folder,
- a Dropbox folder,
- a Sharepoint folder,
- email attachments,
- documents incoming over a Kafka topic,
- or any other stream.
We first read the directory that stores PDFs and then extract the unformatted text from the PDFs using the callable class UnstructuredParser provided in Pathway LLM xpack.
To read the PDFs, we use the pw.io.fs connector, which is used to read documents from the local file system. We need to pass the path and the format, which is binary in our case, since the text is stored in PDFs and Word documents. An additional mode argument exists for whether we use the static or streaming mode. This parameter is not set here as streaming is the default value.
Once the PDFs are loaded, we extract the text using UnstructuredParser(). Since it returns a list of tuples with text and metadata, we filter the metadata out. This is done using strip_metadata, a Pathway user-defined function (UDF). In Pathway, you can easily define a UDF using the @pw.udf decorator.
@pw.udf
def strip_metadata(docs: list[tuple[str, dict]]) -> list[str]:
return [doc[0] for doc in docs]
files = pw.io.fs.read(
data_dir,
format="binary",
)
parser = UnstructuredParser()
unstructured_documents = files.select(texts=parser(pw.this.data))
unstructured_documents = unstructured_documents.select(texts=strip_metadata(pw.this.texts))
2. Extracting information from the text
Now that we have the text stored in a Pathway table, we use structure_on_the_fly to extract the relevant information from the text.
Let's see how structure_on_the_fly works.
First, we need to build the prompt sent to the LLM model. This is done using build_prompt_structure, a Pathway user-defined function (UDF).
The prompt is a long string that gives the LLM all the information it needs to extract from the raw text.
For the LLM, we use OpenAI's GPT-3.5 Turbo model. Pathway supports other LLM models such as HuggingFace, LiteLLM or similar.
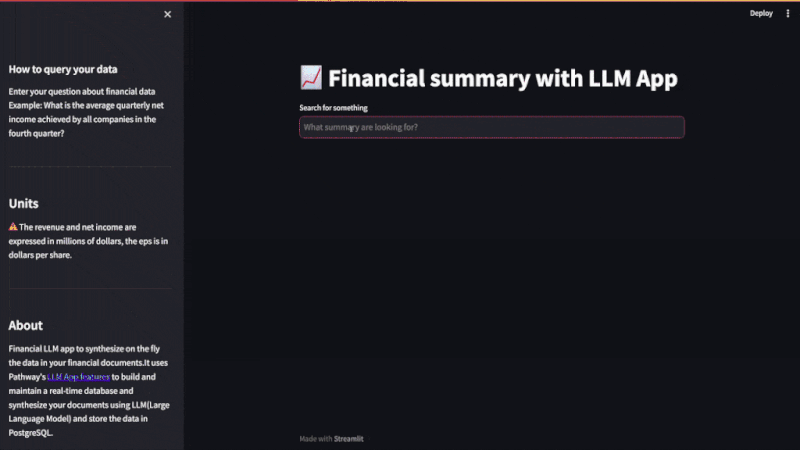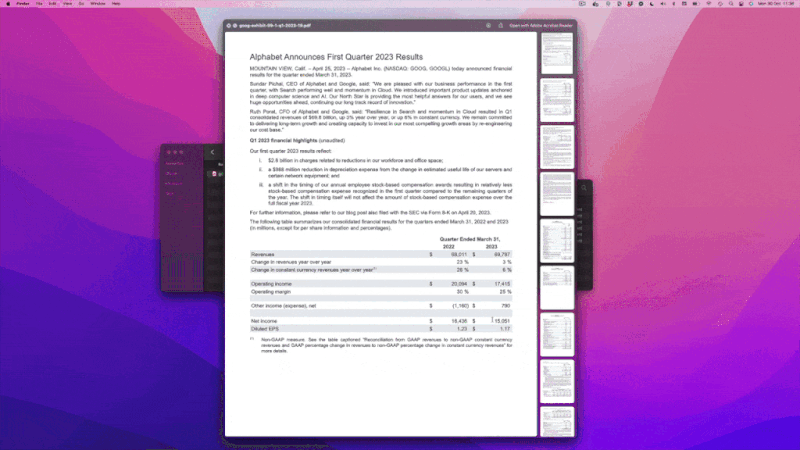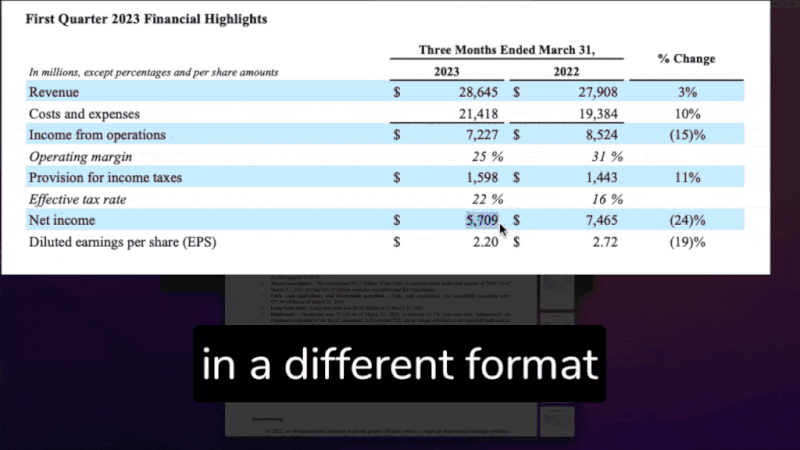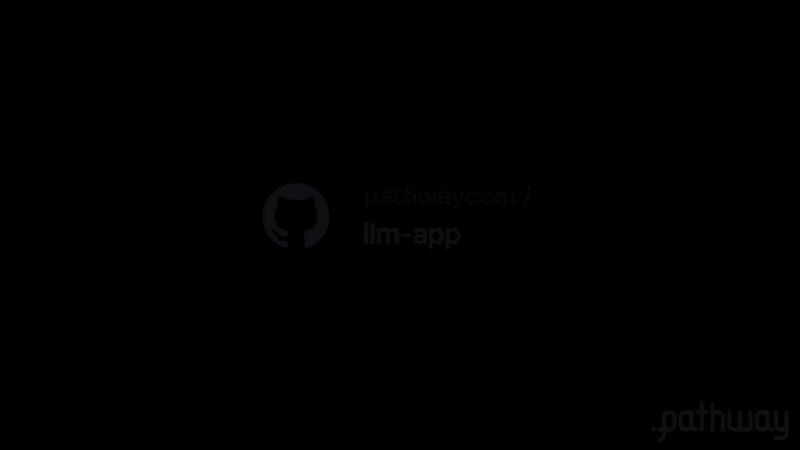
In particular, this is where the relevant data to be extracted is defined: in our example, we displayed the PostgreSQL table schema.
columns are from {postresql_table} table whose schema is:
| Column Name | Data Type |
|-------------------|------------|
| company_symbol | str |
| year | int |
| quarter | str |
| revenue_md | float |
| eps | float |
| net_income_md | float |
The associated Pathway table has the following schema:
class FinancialStatementSchema(pw.Schema):
company_symbol: str
year: int
quarter: str
revenue_md: float
eps: float
net_income_md: float
The llm-app already does all of this in build_prompt_structure, so to obtain the prompt, we simply have to call it:
prompt = documents.select(prompt=build_prompt_structure(pw.this.texts))
pw.this.texts refers to the texts field in the documents table that stores the PDFs.
We need to call the LLM to extract the data with the prompt we prepared.
Pathway is compatible with several models, including OpenAI and any open-source models hosted on the HuggingFace and LiteLLM. We will go with the OpenAI GPT-3.5 Turbo model for ease of use and accuracy. We initialize the model with OpenAIChat provided in the LLM xpack. To fit the prompt into format required by OpenAI API we use a function prompt_chat_single_qa from LLM xpack.
We call OpenAI API with our prompt, and LLM returns its answer as text. This result is stored in the result column of the responses table.
model = OpenAIChat(
api_key=api_key,
model=model_locator,
temperature=temperature,
max_tokens=max_tokens,
retry_strategy=pw.asynchronous.FixedDelayRetryStrategy(),
cache_strategy=pw.asynchronous.DefaultCache(),
)
responses = prompt.select(
result=model(prompt_chat_single_qa(pw.this.prompt)),
)
The LLM returns raw text as output. If the result is correct, we can parse it as a JSON. After parsing items into a dictionary, we put the obtained values in a list.
This parsing step is done using the parse_str_to_list UDF provided by the llm-app.
Now, we have a list in the values column of the responses table.
We need to unpack all these values into table columns so that instead of a single column of values with [foo, bar, ...], we will have a table such as
col1 col2 ...
_____________
foo bar ...
This is easily doable in Pathway using the Pathway unpack_col method:
result = unpack_col(responses.values, *sorted(FinancialStatementSchema.keys()))
The column names are the keys of the schema FinancialStatementSchema. The column names are sorted to be sure to obtain a deterministic order.
Note: for those who are not experienced with Python, * in front unpacks the keys.
Finally, we cast the numerical columns to their respective types, such as float:
result = result.with_columns(
eps=pw.apply(float, pw.this.eps),
net_income_md=pw.apply(float, pw.this.net_income_md),
revenue_md=pw.apply(float, pw.this.revenue_md),
)
.with_columns() is used to keep all the columns except eps, net_income_md and revenue_md without any modification.
The other columns are cast using apply, which applies a function and enforces a proper type.
3. Inserting the data into PostgreSQL
Pathway comes with a connector to PostgreSQL out of the box. You can view the complete list of connectors in our I/O API docs. Using the connector, inserting the table in PostgreSQL is very easy:
pw.io.postgres.write(structured_table, postgreSQL_settings, postresql_table)
structured_table is the resulting Pathway table we want to output to PostgreSQL; postresql_table is the table name in the PostgreSQL database; and settings is a dictionary with the PostgreSQL parameters such as host or the port.
You can learn more about the PostgreSQL output connector in the documentation.
Running the project
Now that all the pipeline is ready, you have to run it!
Don't forget to add pw.run(); otherwise, your pipeline will be built, but no data will be ingested.
To run it, a simple python app.py should do the trick. It is also possible to run with Docker, see the instructions on GitHub for details.
This pipeline will automatically read your PDF files, extract the most relevant information, and store it in a PostgreSQL table.
You can learn more about how this works or how to query the PostgreSQL table using natural language by looking at the source on our GitHub repository
Join our Discord community and dive into discussions on tricks and tips for mastering Retrieval Augmented Generation